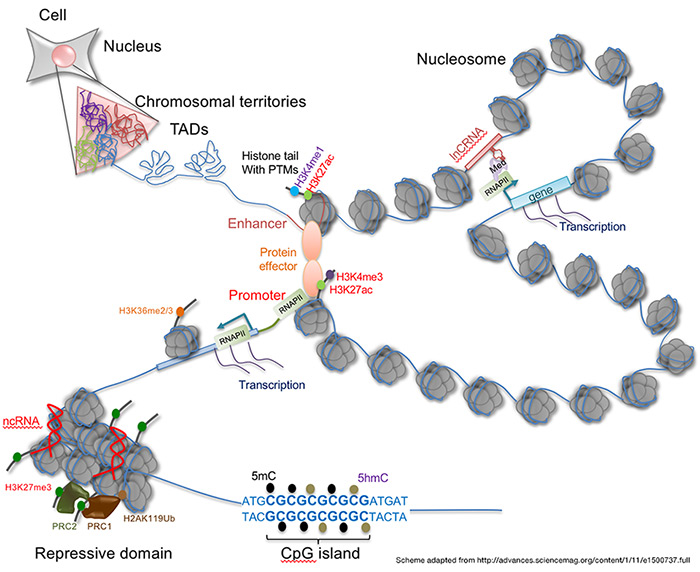
Overview
It is well established that changes in cellular phenotypes that occur during development are highly dependent on the patterning and distribution of epigenetic marks in the genome. Indeed, the major focus of mechanistic epigenetic studies has centered around the interplay between histone or DNA modifying proteins and transcription factors. These studies have been immensely fruitful and yielded many fundamental insights into gene regulation in health and disease. Yet the enormous length of the genome and the requirement for large sets of genes to be regulated and transcribed in a coordinated manner poses significant energetic and physical challenges to cells. It is thus not surprising that an additional critical level of control of the epigenome is conferred through the three dimensional structure of chromosomes and their organization within the nucleus.
Current research focus:
Emerging studies from multiple labs indicate that disruptions in chromosome organization may cause diseases through transcriptional reprogramming. We have extended our studies into this important and exciting area and will use the tools we have developed to determine the impact of nuclear organization in altering gene regulation with a specific focus on developmental regulation and cancer. Projects include:
- Mutations in architectural proteins and their impact on chromosome organization and gene regulation
- Alterations in chromatin modifications and their contribution to changes in chromosome landscape and gene regulation
- High-throughput non-coding CRISPR screens for identifying functional regulatory elements
- The impact of endogenous retroviruses on nuclear organization and gene expression.
Achievements
The Skok Lab’s work has been at the forefront of studies demonstrating that nuclear organization and long-range chromatin interactions play essential roles in gene regulation. Our initial studies focused on lymphocyte development and the control of V(D)J recombination, a process that is important for generating a diverse repertoire of B and T cell receptors that can recognize and combat a wide variety of foreign antigens as part of the adaptive immune response. Using FISH combined with confocal microscopy, the Skok Lab was among the first to demonstrate that chromatin can form dynamic loops. We showed that formation of reversible intra-locus loops in the immunoglobulin (Igh and Igk) and T cell receptor (Tcra and Tcrb) loci have a profound impact on the ability of B and T cells to generate receptor diversity. Furthermore, we were among the first labs to reveal that dynamic reorganization of genes in different nuclear compartments plays important roles in developmental regulation. These studies opened up a whole new field in lymphocyte biology and provided important insight into gene regulation as a whole. Examples of the work pioneered by the Skok Lab lab include the findings that:
- lineage specificity involves maintaining the immunoglobulin (Ig) loci at the nuclear periphery in T cells;
- relocation to the center of the nucleus in early B cell development is associated with opening the locus for accessibility to the recombinase enzyme (RAG) and the initiation of V(D)J recombination;
- reversible dynamic alterations in locus conformation—namely, contraction/intra-locus looping—regulate ordered rearrangement and accessibility of distal Variable (V) gene segments within the Ig and Tcr loci, which is important for maximizing receptor diversity;
- once Igh recombination occurs, interactions with Igk cause Igh to revert to an extended conformation (restricting further recombination) and reposition it to repressive pericentromeric heterochromatin;
- locus decontraction and repositioning to pericentromeric heterochromatin are important for allelic exclusion, which ensures that each lymphocyte expresses a receptor of a single specificity;
- homologous pairing of Ig alleles during V(D)J recombination contributes to allelic exclusion and protects genome stability;
- differential positioning of Ig alleles maintains monoallelic expression in mature B cells;
- signaling molecules and transcription factors regulate locus conformation and nuclear location;
- The recombinase enzyme (RAG) and the DNA repair factor, ATM control RAG activity to limit the number of potential translocation substrates through modulation of 3D conformation (higher order loops) and repositioning of uncleaved loci to repressive pericentromeric heterochromatin;
- chromosomal domains in close proximity to Igh in the nucleus of class switching B cells are predisposed to AID mediated translocations;
- active and Inactive enhancers cooperate to exert localized and long-range control of gene regulation;
- contacts between active enhancers that constitute an Igk super-enhancer in B cells contribute to the transcriptional output of each component;
- 53BP1 plays a damage-independent role that Impacts break order and Igh architecture during class switch recombination, a process that improves antibody effector function.

