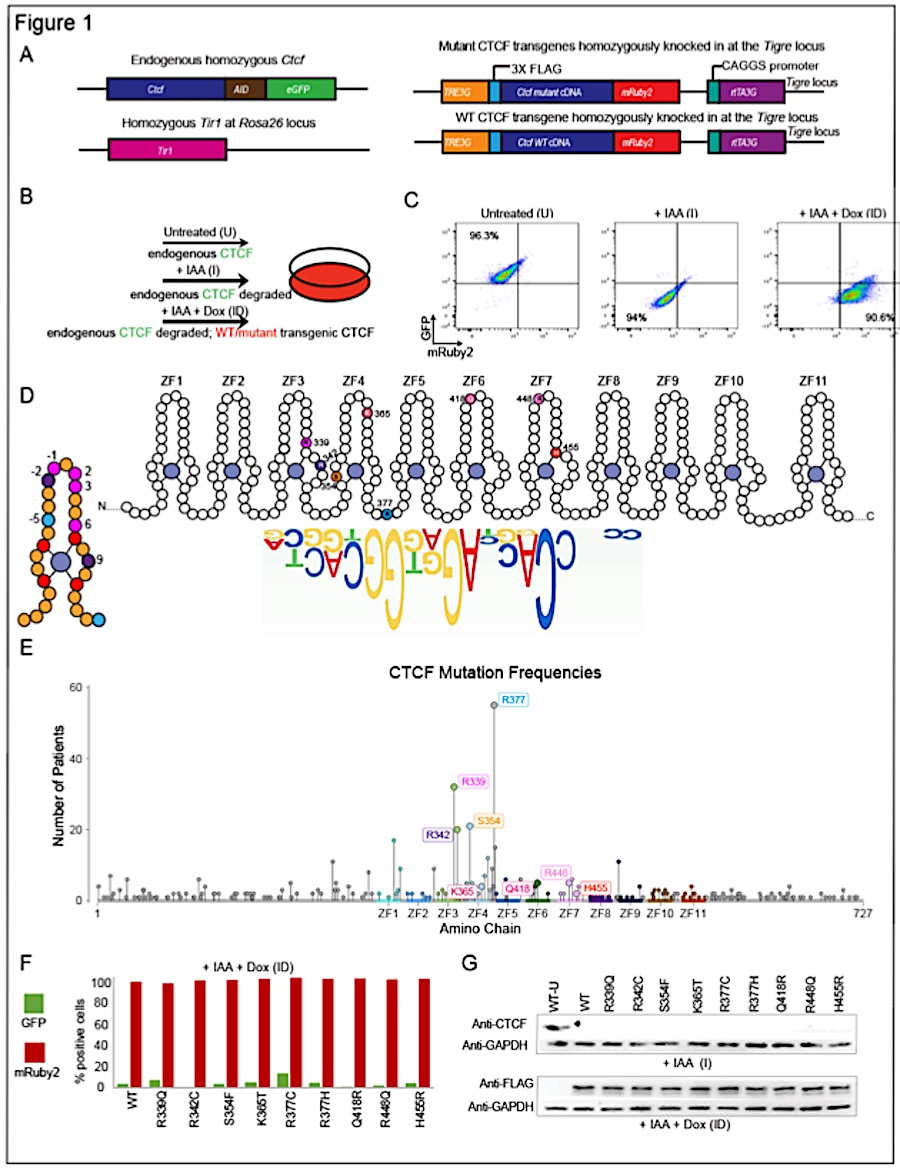
 Brain and cancer associated binding domain mutations provide insight into CTCF’s relationship with chromatin and its ability to act as a chromatin organizer
Brain and cancer associated binding domain mutations provide insight into CTCF’s relationship with chromatin and its ability to act as a chromatin organizer
Catherine Do, Guimei Jiang, Christos C. Katsifis, Domenic N. Narducci, Jie Yang, Giulia Cova, Theodore Sakellaropoulos, Raphael Vidal, Priscillia Lhoumaud, Faye Fara Regis, Nata Kakabadze, Elphege P Nora, Marcus Noyes, Xiaodong Chen, Anders S. Hansen, and Jane A. Skok.
Although only a fraction of CTCF motifs are bound in any cell type, and few occupied sites overlap cohesin, the mechanisms underlying cell-type specific attachment and ability to function as a chromatin organizer remain unknown. To investigate the relationship between CTCF and chromatin we applied a combination of imaging, structural and molecular approaches, using a series of brain and cancer associated CTCF mutations that act as CTCF perturbations. We demonstrate that binding and the functional impact of WT and mutant CTCF depend not only on the unique binding properties of each protein, but also on the genomic context of bound sites and enrichment of motifs for expressed TFs abutting these sites. Our studies also highlight the reciprocal relationship between CTCF and chromatin, demonstrating that the unique binding properties of WT and mutant proteins have a distinct impact on accessibility, TF binding, cohesin overlap, chromatin interactivity and gene expression programs, providing insight into their cancer and brain related effects.
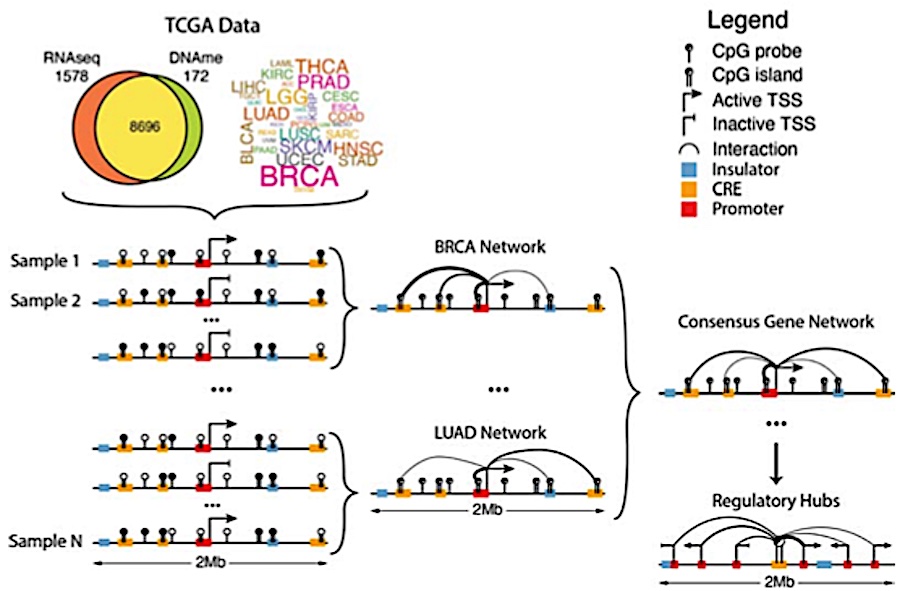 MethNet: a robust approach to identify regulatory hubs and their distal targets in cancer
MethNet: a robust approach to identify regulatory hubs and their distal targets in cancer
Theodore Sakellaropoulos, Catherine Do, Guimei Jiang, Giulia Cova, Peter Meyn, Dacia Dimartino, Sitharam Ramaswami, Adriana Heguy, Aristotelis Tsirigos, and Jane A Skok.
Aberrations in the capacity of DNA/chromatin modifiers and transcription factors to bind non-coding regions can lead to changes in gene regulation and impact disease phenotypes. However, identifying distal regulatory elements and connecting them with their target genes remains challenging. Here, we present MethNet, a pipeline that integrates large-scale DNA methylation and gene expression data across multiple cancers, to uncover novel cis regulatory elements (CREs) in a 1Mb region around every promoter in the genome. MethNet identifies clusters of highly ranked CREs, referred to as ‘hubs’, which contribute to the regulation of multiple genes and significantly affect patient survival. Promoter-capture Hi-C confirmed that highly ranked associations involve physical interactions between CREs and their gene targets, and CRISPRi based scRNA Perturb-seq validated the functional impact of CREs. Thus, MethNet-identified CREs represent a valuable resource for unraveling complex mechanisms underlying gene expression, and for prioritizing the verification of predicted non-coding disease hotspots.
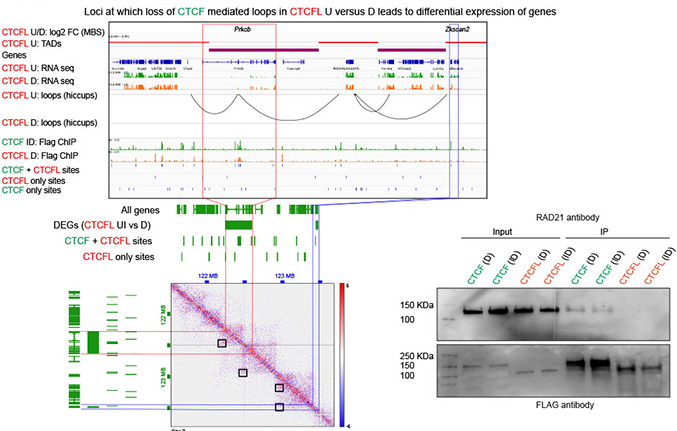 Defining the relative and combined contribution of CTCF and CTCFL to genomic regulation
Defining the relative and combined contribution of CTCF and CTCFL to genomic regulation
Mayilaadumveettil Nishana, Caryn Ha, Javier Rodriguez-Hernaez, Ali Ranjbaran, Erica Chio, Elphege P. Nora, Sana B. Badri, Andreas Kloetgen, Benoit G. Bruneau, Aristotelis Tsirigos and Jane A. Skok.
Ubiquitously expressed CTCF is involved in numerous cellular functions, such as organizing chromatin into TAD structures. In contrast, its paralog, CTCFL, is normally only present in the testis. However, it is also aberrantly expressed in many cancers. While it is known that shared and unique zinc finger sequences in CTCF and CTCFL enable CTCFL to bind competitively to a subset of CTCF binding sites as well as its own unique locations, the impact of CTCFL on chromosome organization and gene expression has not been comprehensively analyzed in the context of CTCF function. Using an inducible complementation system, we analyze the impact of expressing CTCFL and CTCF-CTCFL chimeric proteins in the presence or absence of endogenous CTCF to clarify the relative and combined contribution of CTCF and CTCFL to chromosome organization and transcription. We demonstrate that the N terminus of CTCF interacts with cohesin which explains the requirement for convergent CTCF binding sites in loop formation. By analyzing CTCF and CTCFL binding in tandem, we identify phenotypically distinct sites with respect to motifs, targeting to promoter/intronic intergenic regions and chromatin folding. Finally, we reveal that the N, C, and zinc finger terminal domains play unique roles in targeting each paralog to distinct binding sites to regulate transcription, chromatin looping, and insulation. This study clarifies the unique and combined contribution of CTCF and CTCFL to chromosome organization and transcription, with direct implications for understanding how their co-expression deregulates transcription in cancer.
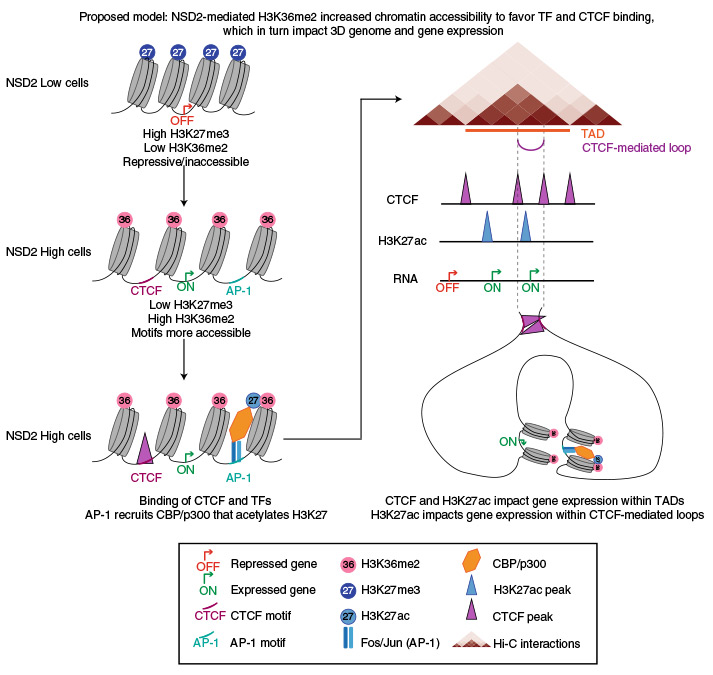 NSD2 overexpression drives clustered chromatin and transcriptional changes in a subset of insulated domains
NSD2 overexpression drives clustered chromatin and transcriptional changes in a subset of insulated domains
Priscillia Lhoumaud,Sana Badri,Javier Rodriguez-Hernaez,Theodore Sakellaropoulos,Gunjan Sethia,Andreas Kloetgen,MacIntosh Cornwell,Sourya Bhattacharyya,Ferhat Ay,Richard Bonneau,Aristotelis Tsirigos & Jane A. Skok
CTCF and cohesin play a key role in organizing chromatin into topologically associating domain (TAD) structures. Disruption of a single CTCF binding site is sufficient to change chromosomal interactions leading to alterations in chromatin modifications and gene regulation. However, the extent to which alterations in chromatin modifications can disrupt 3D chromosome organization leading to transcriptional changes is unknown. In multiple myeloma, a 4;14 translocation induces overexpression of the histone methyltransferase, NSD2, resulting in expansion of H3K36me2 and shrinkage of antagonistic H3K27me3 domains. Using isogenic cell lines producing high and low levels of NSD2, here we find oncogene activation is linked to alterations in H3K27ac and CTCF within H3K36me2 enriched chromatin. A logistic regression model reveals that differentially expressed genes are significantly enriched within the same insulated domain as altered H3K27ac and CTCF peaks. These results identify a bidirectional relationship between 2D chromatin and 3D genome organization in gene regulation.
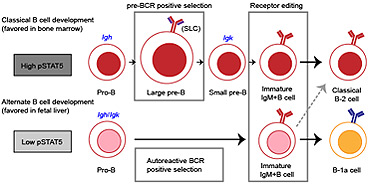 B-1a cells acquire their unique characteristics by bypassing the pre-BCR selection stage
B-1a cells acquire their unique characteristics by bypassing the pre-BCR selection stage
Jason B. Wong, Susannah L. Hewitt, Lynn M. Heltemes-Harris, Malay Mandal, Kristen Johnson, Klaus Rajewsky, Sergei B. Koralov, Marcus R. Clark, Michael A. Farrar & Jane A. Skok
B-1a cells are long-lived, self-renewing innate-like B cells that predominantly inhabit the peritoneal and pleural cavities. In contrast to conventional B-2 cells, B-1a cells have a receptor repertoire that is biased towards bacterial and self-antigens, promoting a rapid response to infection and clearing of apoptotic cells. Although B-1a cells are known to primarily originate from fetal tissues, the mechanisms by which they arise has been a topic of debate for many years. Here we show that in the fetal liver versus bone marrow environment, reduced IL-7R/STAT5 levels promote immunoglobulin kappa gene recombination at the early pro-B cell stage. As a result, differentiating B cells can directly generate a mature B cell receptor (BCR) and bypass the requirement for a pre-BCR and pairing with surrogate light chain. This ‘alternate pathway’ of development enables the production of B cells with self-reactive, skewed specificity receptors that are peculiar to the B-1a compartment. Together our findings connect seemingly opposing lineage and selection models of B-1a cell development and explain how these cells acquire their unique properties.
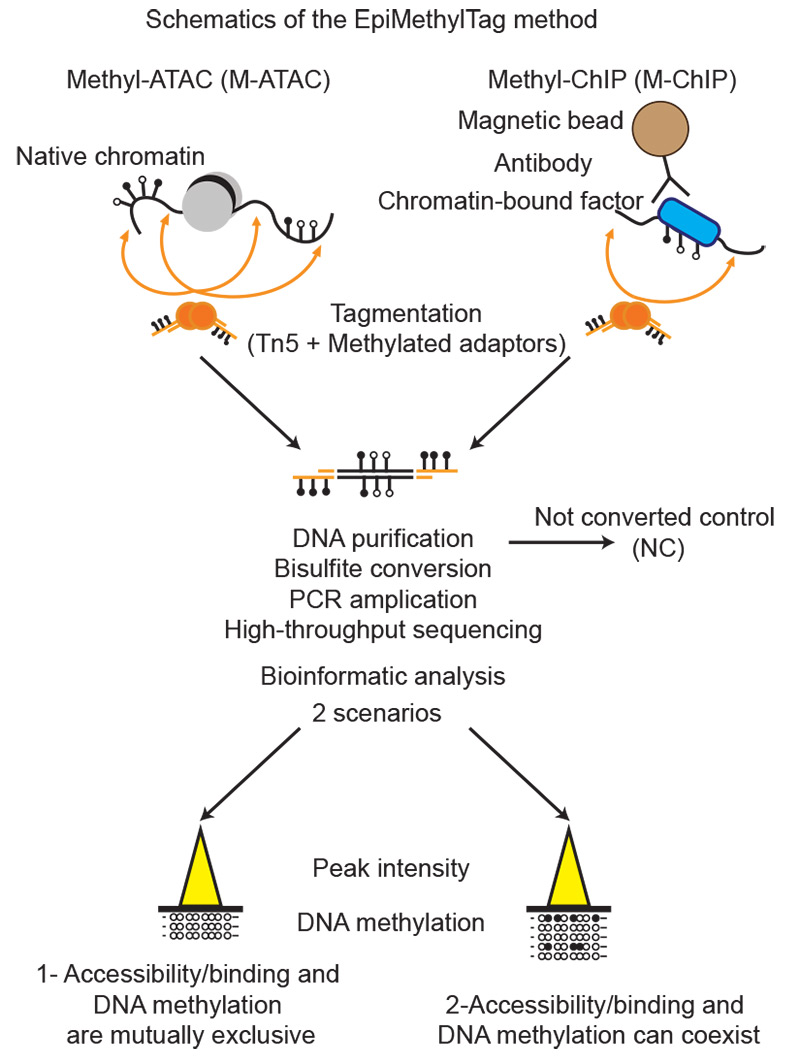 EpiMethylTag: simultaneous detection of ATAC-seq or ChIP-seq signals with DNA methylation
EpiMethylTag: simultaneous detection of ATAC-seq or ChIP-seq signals with DNA methylation
Priscillia Lhoumaud, Gunjan Sethia, Franco Izzo, Theodore Sakellaropoulos, Valentina Snetkova, Simon Vidal, Sana Badri, Macintosh Cornwell, Dafne Campigli Di Giammartino, Kyu-Tae Kim, Effie Apostolou, Matthias Stadtfeld, Dan Avi Landau & Jane Skok
Activation of regulatory elements is thought to be inversely correlated with DNA methylation levels. However, it is difficult to determine whether DNA methylation is compatible with chromatin accessibility or transcription factor (TF) binding if assays are performed separately. We developed a fast, low-input, low sequencing depth method, EpiMethylTag, that combines ATAC-seq or ChIP-seq (M-ATAC or M-ChIP) with bisulfite conversion, to simultaneously examine accessibility/TF binding and methylation on the same DNA. Here we demonstrate that EpiMethylTag can be used to study the functional interplay between chromatin accessibility and TF binding (CTCF and KLF4) at methylated sites.
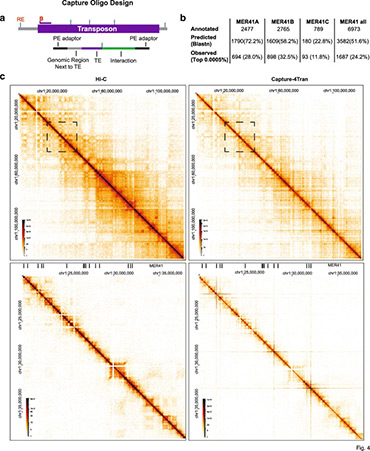 Analysis of 3D genomic interactions identifies candidate host genes that transposable elements potentially regulate
Analysis of 3D genomic interactions identifies candidate host genes that transposable elements potentially regulate
Ramya Raviram, Pedro P. Rocha, Vincent M. Luo, Emily Swanzey,Emily R. Miraldi, Edward B. Chuong, Cédric Feschotte, Richard Bonneau, and Jane A. Skok
The organization of chromatin in the nucleus plays an essential role in gene regulation. About half of the mammalian genome comprises transposable elements. Given their repetitive nature, reads associated with these elements are generally discarded or randomly distributed among elements of the same type in genome-wide analyses. Thus, it is challenging to identify the activities and properties of individual transposons. As a result, we only have a partial understanding of how transposons contribute to chromatin folding and how they impact gene regulation.
Using PCR and Capture-based chromosome conformation capture (3C) approaches, collectively called 4Tran, we take advantage of the repetitive nature of transposons to capture interactions from multiple copies of endogenous retrovirus (ERVs) in the human and mouse genomes. With 4Tran-PCR, reads are selectively mapped to unique regions in the genome. This enables the identification of transposable element interaction profiles for individual ERV families and integration events specific to particular genomes. With this approach, we demonstrate that transposons engage in long-range intra-chromosomal interactions guided by the separation of chromosomes into A and B compartments as well as topologically associated domains (TADs). In contrast to 4Tran-PCR, Capture-4Tran can uniquely identify both ends of an interaction that involve retroviral repeat sequences, providing a powerful tool for uncovering the individual transposable element insertions that interact with and potentially regulate target genes.
4Tran provides new insight into the manner in which transposons contribute to chromosome architecture and identifies target genes that transposable elements can potentially control.
Selected Articles
2022
Cell type-specific prediction of 3D chromatin organization enables high-throughput in silico genetic screening.
Jimin Tan, Nina Shenker-Tauris, Javier Rodriguez-Hernaez, Eric Wang, Theodore Sakellaropoulos, Francesco Boccalatte, Palaniraja Thandapani, Jane Skok, Iannis Aifantis, David Fenyo, Bo Xia, Aristotelis Tsirigos
Link to Article
Editorial overview: Understanding the biology of cancer genomes in the context of DNA, RNA, histone marks, and 3D chromatin organization.
Di Croce, Luciano; Skok, Jane.
Current Opinion in Genetics & Development. 2022
Link to Article
Returning to the lab after a career break.
Skok, Jane A.
Nature Reviews. Molecular Cell Biology. 2022
Link to Article
Ontogeny and Vulnerabilities of Drug-Tolerant Persisters in HER2+ Breast Cancer.
Chang, Chewei Anderson; Jen, Jayu; Jiang, Shaowen; Sayad, Azin; Mer, Arvind Singh; Brown, Kevin R; Nixon, Allison M L; Dhabaria, Avantika; Tang, Kwan Ho; Venet, David; Sotiriou, Christos; Deng, Jiehui; Wong, Kwok-Kin; Adams, Sylvia; Meyn, Peter; Heguy, Adriana; Skok, Jane A; Tsirigos, Aristotelis; Ueberheide, Beatrix; Moffat, Jason; Singh, Abhyudai; Haibe-Kains, Benjamin; Khodadadi-Jamayran, Alireza; Neel, Benjamin G.
Cancer Discovery. 2022
Link to Article
The art of chromosome dynamics: an interview with Jane Skok.
Skok, Jane A.
Epigenomics. 2022
Link to Article
CRISPR and biochemical screens identify MAZ as a cofactor in CTCF-mediated insulation at Hox clusters.
Ortabozkoyun H, Huang PY, Cho H, Narendra V, LeRoy G, Gonzalez-Buendia E, Skok, Jane A, Tsirigos A, Mazzoni EO, Reinberg D.
Nat Genet. 2022
Link to Article
2021
Ontogeny and Vulnerabilities of Drug-Tolerant Persisters in HER2+ Breast Cancer.
Chang CA, Jen J, Jiang S, Sayad A, Mer AS, Brown KR, Nixon AML, Dhabaria A, Tang KH, Venet D, Sotiriou C, Deng J, Wong KK, Adams S, Meyn P, Heguy A, Skok JA, Tsirigos A, Ueberheide B, Moffat J, Singh A, Haibe-Kains B, Khodadadi-Jamayran A, Neel BG.
Cancer Discov. 2022
Link to Article
Simultaneous Tagmentation-Based Detection of ChIP/ATAC Signal with Bisulfite Sequencing.
Lhoumaud P, Skok J.
Methods Mol Biol. 2021
Link to Article
Editorial: From chromatin to dynamic loops and liquid-like phases: New views on the cell nucleus.
Skok J, Gerlich DW.
Current opinion in cell biology. 2021
Link to Article
2020
Somatic Focal Copy Number Gains of Noncoding Regions of Receptor Tyrosine Kinase Genes in Treatment-Resistant Epilepsy.
Vasudevaraja, Varshini; Rodriguez, Javier Hernaez; Pelorosso, Cristiana; Zhu, Kaicen; Buccoliero, Anna Maria; Onozato, Maristela; Mohamed, Hussein; Serrano, Jonathan; Tredwin, Lily; Garonzi, Marianna; Forcato, Claudio; Zeck, Briana; Ramaswami, Sitharam; Stafford, James; Faustin, Arline; Friedman, Daniel; Hidalgo, Eveline Teresa; Zagzag, David; Skok, Jane; Heguy, Adriana; Chiriboga, Luis; Conti, Valerio; Guerrini, Renzo; Iafrate, A John; Devinsky, Orrin; Tsirigos, Aristotelis; Golfinos, John G; Snuderl, Matija.
Journal of neuropathology & experimental neurology. 2020
Link to Article
Scaffold association factor B (SAFB) is required for expression of prenyltransferases and RAS membrane association
Zhou, Mo; Kuruvilla, Leena; Shi, Xiarong; Viviano, Stephen; Ahearn, Ian M; Amendola, Caroline R; Su, Wenjuan; Badri, Sana; Mahaffey, James; Fehrenbacher, Nicole; Skok, Jane; Schlessinger, Joseph; Turk, Benjamin E; Calderwood, David A; Philips, Mark R.
Proceedings of the National Academy of Sciences of the United States of America (PNAS). 2020.
Link to Article
The Ig heavy chain protein but not its message controls early B cell development
Muhammad Assad Aslam, Mir Farshid Alemdehy, Bingtao Hao, Peter H. L. Krijger, Colin E. J. Pritchard, Iris de Rink, Fitriari Izzatunnisa Muhaimin, Ika Nurzijah, Martijn van Baalen, Ron M. Kerkhoven, Paul C. M. van den Berk, Jane A. Skok, and Heinz Jacobs.
PNAS first published November 23, 2020
Link to Article
The novel lncRNA BlackMamba controls the neoplastic phenotype of ALK− anaplastic large cell lymphoma by regulating the DNA helicase HELLS
Valentina Fragliasso, Akanksha Verma, Gloria Manzotti, Annalisa Tameni, Rohan Bareja, Tayla B. Heavican Javeed Iqbal5, Rui Wang, Danilo Fiore, Valentina Mularoni, Wing C. Chan, Priscillia Lhoumaud, Jane Skok, Eleonora Zanetti, Francesco Merli, Alessia Ciarrocchi Oliver Elemento2 Giorgio Inghirami .
Leukemia volume 34, pages2964–2980(2020)
Link to Article
Context-Dependent Requirement of Euchromatic Histone Methyltransferase Activity during Reprogramming to Pluripotency
Vidal, Simon E; Polyzos, Alexander; Chatterjee, Kaushiki; Ee, Ly-Sha; Swanzey, Emily; Morales-Valencia, Jorge; Wang, Hongsu; Parikh, Chaitanya N; Amlani, Bhishma; Tu, Shengjiang; Gong, Yixiao; Snetkova, Valentina; Skok, Jane A; Tsirigos, Aristotelis; Kim, Sangyong; Apostolou, Effie; Stadtfeld, Matthias.
Stem cell reports. 2020.
Link to Article
Defining the relative and combined contribution of CTCF and CTCFL to genomic regulation.
Mayilaadumveettil Nishana, Caryn Ha, Javier Rodriguez-Hernaez, Ali Ranjbaran, Erica Chio, Elphege P. Nora, Sana B. Badri, Andreas Kloetgen, Benoit G. Bruneau, Aristotelis Tsirigos and Jane A. Skok.
Genome Biology, 2020 May 11; 21:108.
CTCF and CTCFL in cancer.
Debaugny RE, Skok JA.
Curr Opin Genet Dev. 2020 Apr 22;61:44‐52. doi:10.1016/j.gde.2020.02.021
DNA methylation disruption reshapes the hematopoietic differentiation landscape.
Izzo F, Lee SC, Poran A, Chaligne R, Gaiti F, Gross B, Murali RR, Deochand SD, Ang C, Jones PW, Nam AS, Kim KT, Kothen-Hill S, Schulman RC, Ki M, Lhoumaud P, Skok JA, Viny AD, Levine RL, Kenigsberg E, Abdel-Wahab O, Landau DA.
Nat Genet. 2020 Apr;52(4):378-387. doi: 10.1038/s41588-020-0595-4. Epub 2020 Mar 23. PubMed PMID: 32203468.
Link to Article
The novel lncRNA BlackMamba controls the neoplastic phenotype of ALK- anaplastic large cell lymphoma by regulating the DNA helicase HELLS.
Fragliasso V, Verma A, Manzotti G, Tameni A, Bareja R, Heavican TB, Iqbal J, Wang R, Fiore D, Mularoni V, Chan WC, Lhoumaud P, Skok J, Zanetti E, Merli F, Ciarrocchi A, Elemento O, Inghirami G.
Leukemia. 2020 Mar 2;. doi: 10.1038/s41375-020-0754-8. [Epub ahead of print] PubMed PMID: 32123306.
Link to Article
2019
Low grade astrocytoma mutations cooperate to disrupt SOX2 genomic architecture and block differentiation via previously unidentified enhancer elements [Meeting Abstract]
Bready, D; Modrek, A; Guerrera, A; Frenster, J; Skok, J; Placantonakis, D.
Neuro-oncology. 2019.
Link to Article
EpiMethylTag: simultaneous detection of ATAC-seq or ChIP-seq signals with DNA methylation
Lhoumaud P, Sethia G, Izzo F, Sakellaropoulos T, Snetkova V, Vidal S, Badri S, Cornwell M, Di Giammartino DC, Kim KT, Apostolou E, Stadtfeld M, Landau DA, Skok J. Genome Biol. 2019 Nov 21;20(1):248. doi: 10.1186/s13059-019-1853-6. PubMed PMID: 31752933.
Link to Article
NSD2 overexpression drives clustered chromatin and transcriptional changes in a subset of insulated domains
Lhoumaud P, Badri S, Rodriguez-Hernaez J, Sakellaropoulos T, Sethia G, Kloetgen A, Cornwell M, Bhattacharyya S, Ay F, Bonneau R, Tsirigos A, Skok JA. Nat Commun. 2019 Oct 24;10(1):4843. doi: 10.1038/s41467-019-12811-4. PubMed PMID: 31649247.
Link to Article
B-1a cells acquire their unique characteristics by bypassing the pre-BCR selection stage
Wong JB, Hewitt SL, Heltemes-Harris LM, Mandal M, Johnson K, Rajewsky K, Koralov SB, Clark MR, Farrar MA, Skok JA. Nat Commun2019 Oct 18;10(1):4768. doi: 10.1038/s41467-019-12824-z. PubMed PMID: 31628339; PubMed Central PMCID: PMC6802180.
Link to Article
RNA interactions are essential for CTCF-mediated genome organization
Ricardo Saldaña-Meyer*, Javier Rodriguez-Hernaez, Thelma Escobar, Mayilaadumveettil Nishana, Karina Jácome-López, Elphege P. Nora, Benoit G. Bruneau,Mayra Furlan-Magaril 4, Jane A Skok, and Danny Reinberg*. Journal: Molecular Cell - MOLECULAR-CELL-D-19-00175R3
Link to Article
A B-cell acute lymphoblastic leukemia regulatory network defines novel therapeutic targets in IGH-CRLF2 patients.
Biorxiv. 2019 May.31. Sana Badri, Beth Carella, Priscillia Lhoumaud, Dayanne M Castro, Ramya Raviram, Aaron Watters, Richard Bonneau, Jane A Skok
Link to article
NSD2 overexpression drives clustered chromatin and transcriptional changes in insulated domains.
Biorxiv. 2019 March.24. Priscillia Lhoumaud, Sana Badri, Javier Rodriguez Hernaez, Gunjan Sethia, Andreas Kloetgen, MacIntosh Cornwell, Sourya Bhattacharyya, Ferhat Ay, Richard Bonneau, Aristotelis Tsirigos, Jane A Skok
Link to article
EpiMethylTag simultaneously detects ATAC-seq or ChIP-seq signals with DNA methylation
Biorxiv. 2019 February.14. Priscillia Lhoumaud, Gunjan Sethia, Franco Izzo, Sana Badri, MacIntosh Cornwell, Kyu-Tae Kim, Dan Landau, Jane Skok
Link to article
Impaired Expression of Rearranged Immunoglobulin Genes and Premature p53 Activation Block B Cell Development in BMI1 Null Mice.
Cantor DJ, King B, Blumenberg L, DiMauro T, Aifantis I, Koralov SB, Skok JA, David G. Cell Rep. 2019 Jan 2;26(1):108-118.e4. doi: 10.1016/j.celrep.2018.12.030.
Link to article
2018
Control of B-1a cell development by instructive BCR signaling
Kreslavsky, Taras; Wong, Jason B; Fischer, Maria; Skok, Jane A; Busslinger, Meinrad.
Current opinion in immunology. 2018:51:24-31
Link to Article
Analysis of 3D genomic interactions identifies candidate host genes that transposable elements potentially regulate.
Raviram R, Rocha PP, Luo VM, Swanzey E, Miraldi ER, Chuong EB, Feschotte C, Bonneau R, Skok JA. Genome Biol. 2018 Dec 13;19(1):216. doi: 10.1186/s13059-018-1598-7.
Link to article
Stage-specific epigenetic regulation of CD4 expression by coordinated enhancer elements during T cell development
Nat Commun. 2018 Sep 5;9(1):3594. Issuree, Priya D; Day, Kenneth; Au, Christy; Raviram, Ramya; Zappile, Paul; Skok, Jane A; Xue, Hai-Hui; Myers, Richard M; Littman, Dan R
Link to article
Capturing the Onset of PRC2-Mediated Repressive Domain Formation
Mol Cell. 2018 Jun 21;70(6):1149-1162. Oksuz, Ozgur; Narendra, Varun; Lee, Chul-Hwan; Descostes, Nicolas; LeRoy, Gary; Raviram, Ramya; Blumenberg, Lili; Karch, Kelly; Rocha, Pedro P; Garcia, Benjamin A; Skok, Jane A; Reinberg, Danny
The impact of endogenous retroviruses on nuclear organization and gene expression.
Biorxiv. 2018 Apr. 6. Ramya Raviram, Pedro P Rocha, Vincent Luo, Emily Swanzey, Emily Miraldi, Edward Chuong, Cedric Feschotte, Richard Bonneau, Jane A. Skok.
Link to Article
Enhancer Talk
Epigenomics. 2018 Mar 27. Valentina Snetkova and Jane Skok. doi: 10.2217/epi-2017-0157. PubMed PMID: 29583027.
Link to Article
2017
B-1a cells acquire their unique characteristics by bypassing the pre-BCR selection stage
Biorxiv. 2017 Nov. 13. Jason B Wong, Susannah L Hewitt, Lynn M Heltemes-Harris, Malay Mandal, Kristen Johnson, Klaus Rajewsky, Sergei B Koralov, Marcus R Clark, Michael A Farrar, Jane A. Skok.
Link to Article
Low-Grade Astrocytoma Mutations in IDH1, P53, and ATRX Cooperate to Block Differentiation of Human Neural Stem Cells via Repression of SOX2.
Cell Rep. 2017 Oct. 31; 21(5). Aram S. Modrek, Danielle Golub, Themasap Khan, Devin Bready, Jod Prado, Christopher Bowman, Jingjing Deng, Guoan Zhang, Pedro P. Rocha, Ramya Raviram, Charalampos Lazaris, James M. Stafford, Gary LeRoy, Michael Kader, Joravar Dhaliwal, N. Sumru Bayin, Joshua D. Frenster, Jonathan Serrano, Luis Chiriboga, Rabaa Baitalmal, Gouri Nanjangud, Andrew S. Chi, John G. Golfinos, Jing Wang, Matthias A. Karajannis, Richard A. Bonneau, Danny Reinberg, Aristotelis Tsirigos, David Zagzag, Matija Snuderl, Jane A. Skok, Thomas A. Neubert, Dimitris G. Placantonakis.
Link to Article
The Conserved ATM Kinase RAG2-S365 Phosphorylation Site Limits Cleavage Events in Individual Cells Independent of Any Repair Defect.
Cell Rep. 2017 Oct. 24; 21(4). Susannah L. Hewitt, Jason B. Wong, Ji-Hoon Lee, Mayilaadumveettil Nishana, Hongxi Chen, Marc Coussens, Suzzette M. Arnal, Lili M. Blumenberg, David B. Roth, Tanya T. Paull, Jane A. Skok.
Link to Article
Chromatin Folding and Recombination. Nuclear Architecture and Dynamics.
Valentina Snetkova and Jane A. Skok. 2017 Oct 20;475–489.
Wolf-Hirschhorn Syndrome Candidate 1 Is Necessary for Correct Hematopoietic and B Cell Development.
Cell Rep. 2017 May 23;19(8). Elena Campos-Sanchez, Nerea Deleyto-Seldas, Veronica Dominguez, Enrique Carrillo-de-Santa-Pau, Kiyoe Ura, Pedro P. Rocha, JungHyun Kim, Arafat Aljoufi, Anna Esteve-Codina, Marc Dabad, Marta Gut, Holger Heyn, Yasufumi Kaneda, Keisuke Nimura, Jane A. Skok, Maria Luisa Martinez-Frias, Cesar Cobaleda.
Link to Article
The IgH locus 3’ cis-regulatory super-enhancer co-opts AID for allelic transvection.
Oncotarget. 2017 Feb 21; 8(8). Sandrine Le Noir, Brice Laffleur, Claire Carrion, Armand Garot, Sandrine Lecardeur, Eric Pinaud, Yves Denizot, Jane A. Skok, Michel Cogné.
Link to Article
2016
MED12 Regulates HSC-Specific Enhancers Independently of Mediator Kinase Activity to Control Hematopoiesis.
Cell Stem Cell. 2016 Dec 1; 19(6).Beatriz Aranda-Orgilles, Ricardo Saldaña-Meyer, Eric Wang, Eirini Trompouki, Anne Fassl, Stephanie Lau, Jasper Mullenders, Pedro P. Rocha, Ramya Raviram, María Guillamot, María Sánchez-Díaz, Kun Wang, Clarisse Kayembe, Nan Zhang, Leonela Amoasii, Avik Choudhuri, Jane A. Skok, Markus Schober, Danny Reinberg, Piotr Sicinski, Heinrich Schrewe, Aristotelis Tsirigos, Leonard I. Zon, Iannis Aifantis.
Link to Article
miRNAs Are Essential for the Regulation of the PI3K/AKT/FOXO Pathway and Receptor Editing during B Cell Maturation.
Cell Reports. 2016 Nov 22; 17(9). Maryaline Coffre, David Benhamou, David Rieß, Lili Blumenberg, Valentina Snetkova, Marcus J. Hines, Tirtha Chakraborty, Sofia Bajwa, Kari Jensen, Mark M.W. Chong, Lelise Getu, Gregg J. Silverman, Robert Blelloch, Dan R. Littman, Dinis Calado, Doron Melamed, Jane A. Skok, Klaus Rajewsky, Sergei B. Koralov.
Link to Article
Identification of multi-loci hubs from 4C-seq demonstrates the functional importance of simultaneous interactions.
Nucleic Acids Res. 2016 Oct 14; 44(18). Tingting Jiang, Ramya Raviram, Valentina Snetkova, Pedro P. Rocha, Charlotte Proudhon, Sana Badri, Richard Bonneau, Jane A. Skok*, Yuval Kluger*.
Link to Article
A Damage-Independent Role for 53BP1 that Impacts Break Order and Igh Architecture during Class Switch Recombination.
Cell Reports (2016). Pedro P. Rocha*, Ramya Raviram, Yi Fu, JungHyun Kim, Vincent M. Luo, Arafat Aljoufi, Emily Swanzey, Alessandra Pasquarella, Alessia Balestrini, Emily R. Miraldi, Richard Bonneau, John Petrini, Gunnar Schotta, and Jane A. Skok.
Link to Article
Active and Inactive Enhancers Cooperate to Exert Localized and Long-Range Control of Gene Regulation.
Charlotte Proudhon*, Valentina Snetkova*, Ramya Raviram, Camille Lobry, Sana Badri, Tingting Jiang, Bingtao Hao, Thomas Trimarchi, Yuval Kluger, Iannis Aifantis, Richard Bonneau, Jane A. Skok. Cell Reports. 2016 May 25. pii: S2211-1247(16)30532-0. doi: 10.1016/j.celrep.2016.04.087. [Epub ahead of print] PubMed PMID: 27239026.
Link to Article
CRISPR-dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat-enriched individual loci.
Yi Fu*, Pedro P. Rocha*, Vincent M. Luo, Ramya Raviram, Yan Deng, Esteban O. Mazzoni & Jane A. Skok. Nat Commun. 2016 May 25;7:11707. doi: 10.1038/ncomms11707. PubMed PMID: 27222091.
Link to Article
Mediator facilitates transcriptional activation and dynamic long-range contacts at the IgH locus during class switch recombination.
Anne-Sophie Thomas-Claudepierre, Isabelle Robert, Pedro P. Rocha, Ramya Raviram, Ebe Schiavo, Vincent Heyer, Richard Bonneau, Vincent M. Luo, Janardan K. Reddy, Tilman Borggrefe, Jane A. Skok, and Bernardo Reina-San-Martin. J Exp Med. 2016 Feb 22;213(3):
Link to Article
4C-ker: A Method to Reproducibly Identify Genome-Wide Interactions Captured by 4C-Seq Experiments.
Ramya Raviram, Pedro P. Rocha, Christian L. Müller, Emily R. Miraldi, Sana Badri, Yi Fu, Emily Swanzey, Charlotte Proudhon, Valentina Snetkova, Richard Bonneau, Jane A. Skok . PLoS Comput Biol. 2016 Mar 3;
Link to Article
2015
RAG Off-Target Activity Is in the Loop
Blumenberg, Lili; Skok, Jane A.
Trends in molecular medicine. 2015:21(12):733-735
Link to Article
Transcription. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation
Narendra, Varun; Rocha, Pedro P; An, Disi; Raviram, Ramya; Skok, Jane A; Mazzoni, Esteban O; Reinberg, Danny.
Science. 2015:347(6225):1017-21
Link to Article
VH replacement in primary immunoglobulin repertoire diversification
Sun, Amy; Novobrantseva, Tatiana I; Coffre, Maryaline; Hewitt, Susannah L; Jensen, Kari; Skok, Jane A; Rajewsky, Klaus; Koralov, Sergei B.
Proceedings of the National Academy of Sciences of the United States of America (PNAS). 2015:112(5):E458-66
Link to Article
Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms.
Jasper Mullenders, Beatriz Aranda-Orgilles, Priscillia Lhoumaud, Matthew Keller, Juhee Pae, Kun Wang, Clarisse Kayembe, Pedro P. Rocha, Ramya Raviram, Yixiao Gong, Prem K. Premsrirut, Aristotelis Tsirigos, Richard Bonneau, Jane A. Skok, Luisa Cimmino, Daniela Hoehn, and Iannis Aifantis. J Exp Med. 2015 Oct 5;212(11):
Link to Article
Long-Range Regulation of V(D)J Recombination. Advances in Immunology.
Charlotte Proudhon, Bingtao Hao, Ramya Raviram, Julie Chaumeil, and Jane A. Skok. 2015 Aug 20;123–182.
Link to Article
Breaking TADs: insights into hierarchical genome organization.
Pedro P Rocha, Ramya Raviram, Richard Bonneau and Jane A Skok. Epigenomics. 2015 Jun;
Link to Article
CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation.
Varun Narendra, Pedro P. Rocha, Disi An, Ramya Raviram, Jane A. Skok, Esteban O. Mazzoni, and Danny Reinbergc. Science . 2015 Feb 27;347(6225):
Link to Article
VH replacement in primary immunoglobulin repertoire diversification.
Amy Sun, Tatiana I. Novobrantseva, Maryaline Coffre, Susannah L. Hewitt, Kari Jensen, Jane A. Skok, Klaus Rajewsky, and Sergei B. Koralov. PNAS. 2015 Feb;
Link to Article
2014
Taking a break from the lab: can it really be done?
Jane A. Skok. Trends in Cell Biology. 2014 Dec;
Link to Article
Interpreting 4C-Seq data: how far can we go?
Ramya Raviram, Pedro P Rocha, Richard Bonneau, Jane A Skok. Epigenomics. 2014 Dec;6:455-457.
Link to Article
β-Catenin induces T-cell transformation by promoting genomic instability.
Marei Dose, Akinola Olumide Emmanuel, Julie Chaumeil, Jiangwen Zhang, Tianjiao Sun, Kristine Germar, Katayoun Aghajani, Elizabeth M. Davis, Shilpa Keerthivasan, Andrea L. Bredemeyer, Barry P. Sleckman, Steven T. Rosen, Jane A. Skok, Michelle M. Le Beau, Katia Georgopoulos, and Fotini Gounari. PNAS. 2014 Jan;
Link to Article
2013
Finding the Right Partner in a 3D Genome.
Pedro P. Rocha, Julie Chaumeil, Jane A. Skok. Science. 2013 Dec 13;342:1333-1334.
Link to Article
A new take on V(D)J recombination: transcription driven nuclear and chromatin reorganization in RAG-mediated cleavage.
Julie Chaumeil Jane Skok. Frontiers in Immunology. 2013 Dec 6;
Link to Article
Response to Casellas et al
Pedro P. Rocha, Mariann Micsinai, Yuval Kluger, Jane A. Skok. Molecular Cell. 2013 Aug 8;51:
Link to Article
The RAG2 C-terminus and ATM protect genome integrity by controlling antigen receptor gene cleavage.
Julie Chaumeil, Mariann Micsinai, Panagiotis Ntziachristos, David B. Roth, Iannis Aifantis, Yuval Kluger, Ludovic Deriano & Jane A. Skok. Nature Communications. 2013 Jul 2;4:
Link to Article
The origin of recurrent translocations in recombining lymphocytes: a balance between break frequency and nuclear proximity.
Pedro P Rocha, Jane A Skok. Current Opinion in Cell Biology. 2013 Mar 13;25:Epub 2013 Mar 13.
Link to Article
Combined Immunofluorescence and DNA FISH on 3D-preserved Interphase Nuclei to Study Changes in 3D Nuclear Organization.
Julie Chaumeil, Mariann Micsinai, Jane A. Skok. Journal of Visualized Experiments. 2013 Feb 3;72:e50087.
Link to Article
2012
Close Proximity to Igh Is a Contributing Factor to AID-Mediated Translocations. Molecular Cell.
Pedro P. Rocha, Mariann Micsinai, JungHyun Rachel Kim, Susannah L. Hewitt, Patricia P. Souza, Thomas Trimarchi, Francesco Strino, Fabio Parisi, Yuval Kluger, and Jane A. Skok. 2012 Sep 28;47:873-885. Epub 2012 Aug 8.
Link to Article
IL-7 Functionally Segregates the Pro-B Cell Stage by Regulating Transcription of Recombination Mediators across Cell Cycle. Current Opinion in Immunology.
Johnson K, Chaumeil J, Micsinai M, Wang JM, Ramsey LB, Baracho GV, Rickert RC, Strino F, Kluger Y, Farrar MA, Skok JA. 2012 Jun 15;188:6084-92. Epub 2012 May 11.
Link to Article
The role of CTCF in regulating V(D)J recombination.
Chaumeil J, Skok JA. EMBO Journal. 2012 Apr;24:153-9. Epub 2012 Apr 15.
Link to Article
Equal oportunity for all. EMBO Journal.
Chaumeil J, Skok JA. 2012 Mar 13;31:Cited in Pubmed; PMID 22415367.
Link to Article
2011
RUNX Transcription Factor-Mediated Association of Cd4 and Cd8 Enables Coordinate Gene Regulation.
Collins A, Hewitt SL, Chaumeil J, Sellars ML, Micsinai M, Allinne J, Parisi F, Nora E, Bolland DJ, Corcoran AE, Kluger Y, Bosselut R, Ellmeier W, Chong MMW, Littman DR, and Skok JA. Immunity. 2011 Mar 25;34(3):303-14. doi: 10.1016/j.immuni.2011.03.004.
Link to Article
A Multifunctional Element in the Mouse Ig{kappa} Locus That Specifies Repertoire and Ig Loci Subnuclear Location.
Xiang Y, Zhou X, Hewitt SL, Skok JA, Garrard WT. J Immunol. 2011 Apr 1;186(7):000–000. Epub 2011 Mar 25. Cited in Pubmed; PMID 21441452. doi: 10.4049/jimmunol.1003794.
The RAG2 C-terminus suppresses genomic instability and lymphomagenesis.
Deriano L, Chaumeil J, Coussens M, Multani A, Chou YF, Alekseyenko AV, Chang S, Skok JA, Roth DB. Nature. 2011 Mar 3;471(7336):119-23. Epub 2011 Mar 2. doi: 10.1038/nature09755.
Link to Article
2010
Epigenetic regulation of V(D)J recombination. Essays Biochem.
Johnson K, Chaumeil J, Skok JA. 2010 Sep 20;48(1):221-43. Cited in Pubmed; PMID 20822496. doi: 10.1042/bse0480221.
Chromosome dynamics and the regulation of V(D)J recombination.
Hewitt SL, Chaumeil J, Skok JA. Immunol Rev. 2010 Sep;237(1):43-54. Cited in Pubmed; PMID 20727028. doi: 10.1111/j.1600-065X.2010.00931.x.
It takes two: Communication between homologous alleles preserves genomic stability during V(D)J recombination.
Brandt, V.L., Hewitt, S.L., and Skok, J.A. Nucleus. 2010;1:23-29.
V(D)J recombination: a paradigm for studying chromosome interactions in mammalian cells.
Skok, J. Epigenomics. 2010;2:176-177.
2009
RAG1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci.
Hewitt SL*, Yin B*, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, Farrar MA, Sleckman BP, Schatz DG, Busslinger M, Bassing CH*, Skok JA*. Nat Immunol. 2009 Jun;10(6):655-64. Epub 2009 May 17. Cited in Pubmed; PMID 19448632. doi: 10.1038/ni.1735. PMCID: PMC2693356.
2008
Association between the Igk and Igh immunoglobulin loci mediated by the 3' Igk enhancer induces 'decontraction' of the Igh locus.
Hewitt SL, Farmer D, Marszalek K, Cadera E, Liang HE, Xu Y, Schlissel MS, Skok JA. Nat Immunol. 2008 Apr;9(4):396-404. Epub 2008 Feb 24. Cited in Pubmed; PMID 18297074. doi: 10.1038/ni1567. PMCID: PMC2583163.
Regulation of Immunoglobulin Light-Chain Recombination by the Transcription Factor IRF-4 and the Attenuation of Interleukin-7 Signaling.
Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, Singh H. Immunity. 2008 Mar;28(3):335-45. Epub 2008 Feb 14. Cited in Pubmed; PMID 18280186. doi: 10.1016/j.immuni.2007.12.019.
2007
Dynamic changes in accessibility, nuclear positioning, recombination, and transcription at the Ig kappa locus.
Fitzsimmons S, Bernstein R, Skok JA, Max EE, Shapiro MA. J Immunol. 2007 Oct 15;179:5264-73. Cited in Pubmed; PMID 17911612.
Link to Article
Transcriptional regulation in early B cell development.
Fuxa M, Skok JA. Curr Opin Immunol. 2007 Apr;19(2):129-36. Cited in Pubmed; PMID 17292598. doi: 10.1016/j.coi.2007.02.002.
Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes.
Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Nat Immunol. 2007 Apr;8(4):378-87. Cited in Pubmed; PMID 17334367. doi: 10.1038/ni1448.
2005
The pre-B-cell receptor induces silencing of VpreB and lambda5 transcription.
Parker MJ, Licence S, Erlandsson L, Galler GR, Chakalova L, Osborne CS, Morgan G, Fraser P, Jumaa H, Winkler TH, Skok J, Mårtensson lL. EMBO J. 2005 Nov 16;24(22):3895-905. Cited in Pubmed; PMID 15580273. doi: 10.1038/sj.emboj.7600850. PMCID: PMC1283949.
Epigenetic ontogeny of the Igk locus during B cell development.
Goldmit M*, Ji Y*, Skok J*, Roldan E, Jung S, Cedar H, Bergman Y. Nat Immunol. 2005 Feb;6(2):198-203. Cited in Pubmed; PMID 15619624. doi: 10.1038/ni1154.
Locus 'decontraction' and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene.
Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok JA. Nat Immunol. 2005 Jan;6(1):31-41. Cited in Pubmed; PMID 15580273. doi: 10.1038/ni1150. PMCID: PMC1592471.
2004
Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene.
Fuxa M*, Skok J*, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Genes Dev. 2004 Feb 15;18(4):411-22. Cited in Pubmed; PMID 15004008. doi: 10.1101/gad.291504. PMCID: PMC359395.
2002
Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development.
Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Science. 2002 Apr 5;296(5565):158-62. Cited in Pubmed; PMID 11935030. doi: 10.1126/science.1068768.
2001
Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes.
Skok JA, Brown KE, Azuara V, Caparros ML, Baxter J, Takacs K, Dillon N, Gray D, Perry RP, Merkenschlager M, Fisher AG. Nat Immunol. 2001 Sep;2(9):848-54. Cited in Pubmed; PMID 11526401. doi: 10.1038/ni0901-848.
Rewiring of CD40 is necessary for delivery of rescue signals to B cells in germinal centres and subsequent entry into the memory pool.
Siepmann K, Skok J, van Essen D, Harnett M, Gray D. Immunology. 2001 Mar;102(3):263-72. Cited in Pubmed; PMID 11298824. doi: 10.1046/j.1365-2567.2001.01162.x. PMCID: PMC1783186.
1999
Dendritic cell-derived IL-12 promotes B cell induction of Th2 differentiation: a feedback regulation of Th1 development.
Skok J, Poudrier J, Gray D. J Immunol. 1999 Oct 15;163(8):4284-91. Cited in Pubmed; PMID 10510367.
Link to Article
1981
Distinct genes for fibroblast and serum C1q.
Skok J, Solomon E, Reid KB, Thompson RA. Nature. 1981;292:549–51. Cited in Pubmed; PMID 6973093.

